INPUT
You can submit your sequences using the textbox or the upload button.
The textbox submission is completed in two steps (Figure 1).
Step 1: The input sequence must be in FASTA format. You can upload up to ~100,000 sequences (file size less than 40Mb). For predictions in larger datasets users are encouraged to contact katnastou[at]biol.uoa.gr
Step 2: The prediction is performed by pressing the Submit button.
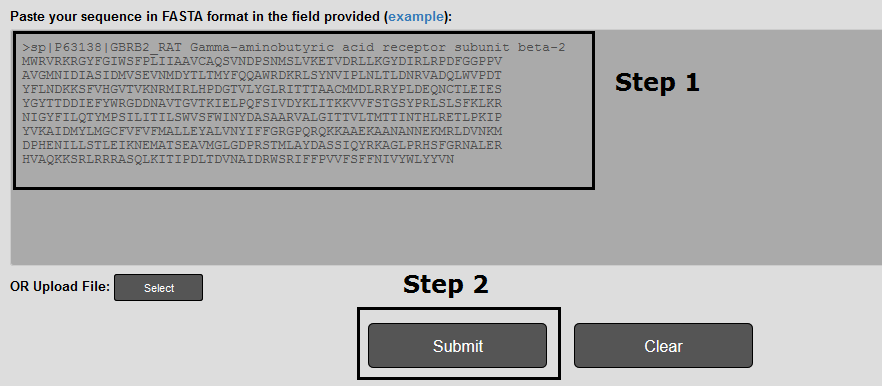
Figure 1. The input page for running LiGIoNs. The textbox submission is completed in two steps, as described above.
You can upload a file with your sequences in two steps (Figure 2).
Step 1: By pressing Select a file selection window pops up.
Step 2: The prediction is performed by pressing the Submit button.
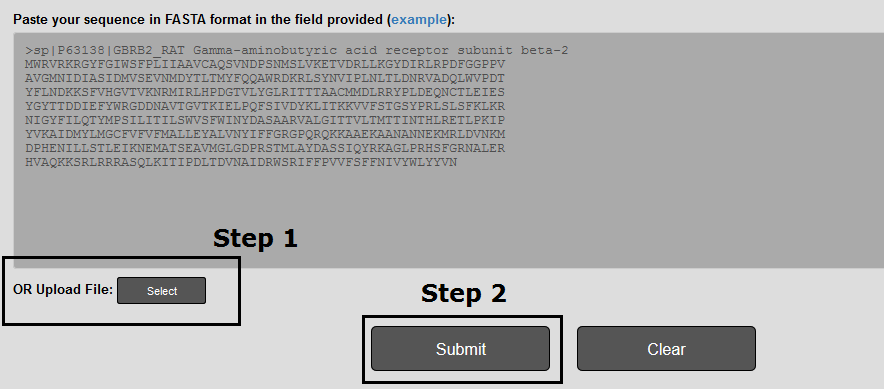
Figure 2. The input page for running LiGIoNs. The upload file submission is completed in two steps, as described above.
RESULTS
In the Results page of LiGIoNs the user can gather information about their submission and download the files with the results.
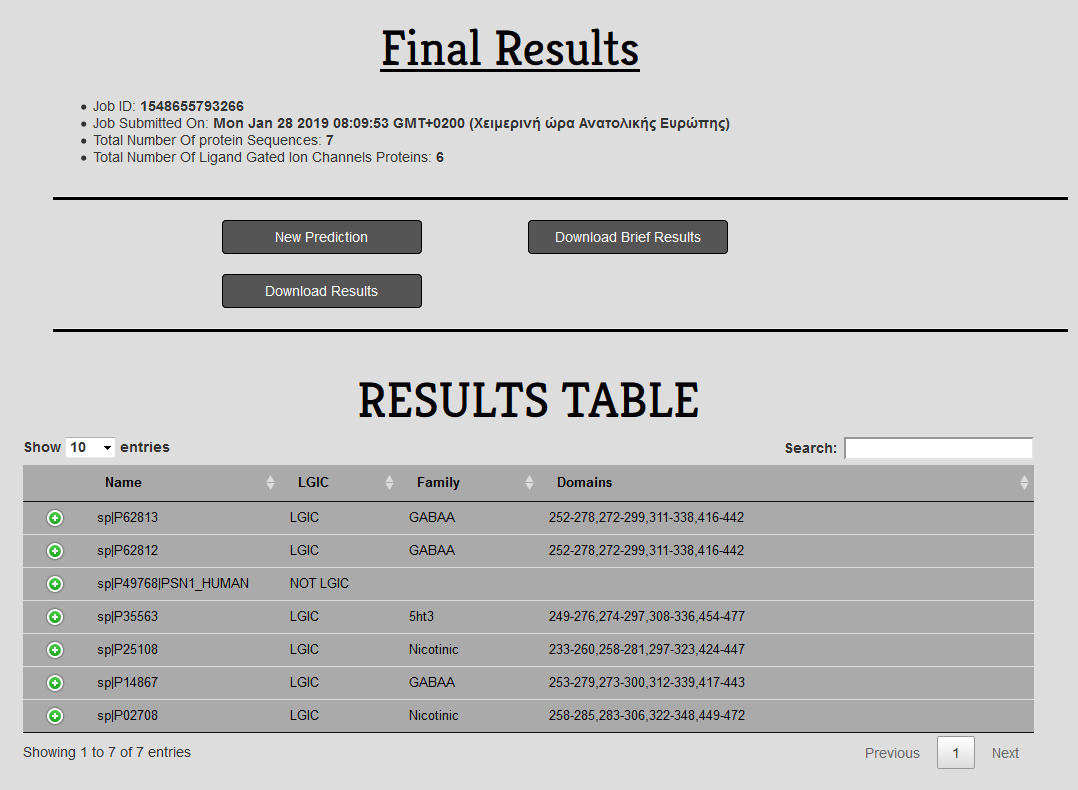
Figure 3. The results page after the submission of seven test sequences.
Submission Information
In the results page the user can gather information regarding their submission. The user can get the Job ID, the Job Submission Date and Time, the Total Number of Submitted Protein Sequences and the Number of Ligand Gated Ion Channels detected by the method

Figure 4. The submission information produced by LiGIoNs for a user query.
Download results
LiGIonS produces two files that a user can download. A results file that contains detailed information about each prediction and a file than contain lists of all predicted Ligand-gated ion channels
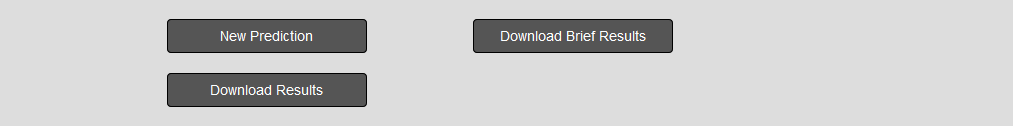
Figure 5. The downloadable results files that are produced after an LiGIoNs submission.
The user can download LiGIoNs prediction results in a text file as shown in Figure 6 by pressing the "Final Results File" button on the results page
In the final results file the user can gather information regarding the predicted LGIC. The first line of the file contains the ID that the user has provided during the submission. The second line starts with RS and contains the results of prediction, if the query protein is an LGIC, in addition to the specific LGIC subfamily. The lines starting with TP contain information about the profiles that 'hit' the protein's sequence. The lines below the line that starts with SQ contain the sequence of the protein. Each entry in the file is separated by other entries with '//end'.
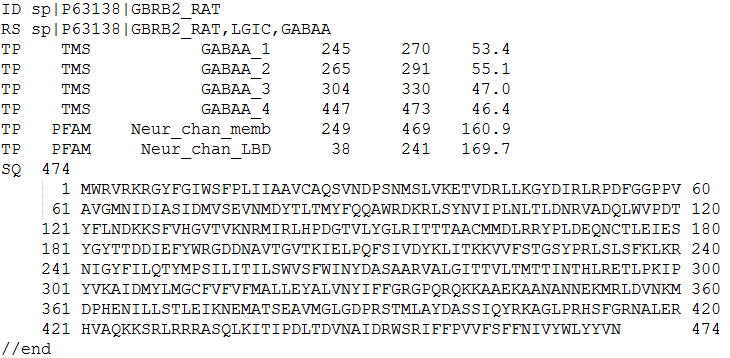
Figure 6.The final text output file of LiGIoNs. Here an example of the Gamma-aminobutyric acid receptor from Rattus norvegicus (Rat) is shown
Note that the text output file is maintained in the server for two weeks and can be accessed through the provided link (using a unique ID number provided for each submission, e.g. 1430043645). All submissions and results are kept confidential and deleted after two weeks.
Webpage Output
After each successful submissions the results are provided to the user as shown in the figure below
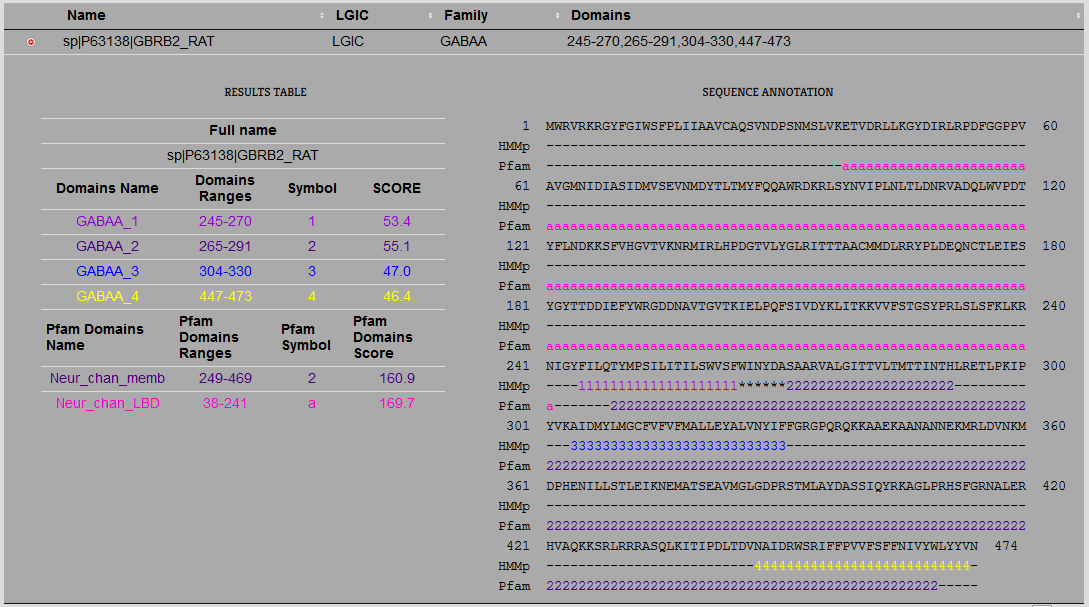
Figure 7. Webpage results after a successful run of LiGIoNs.
Finally, the user can retrieve the results of a prediction by using only the Job ID.
Figure 8. Form to retrieve previous results using a Job ID.