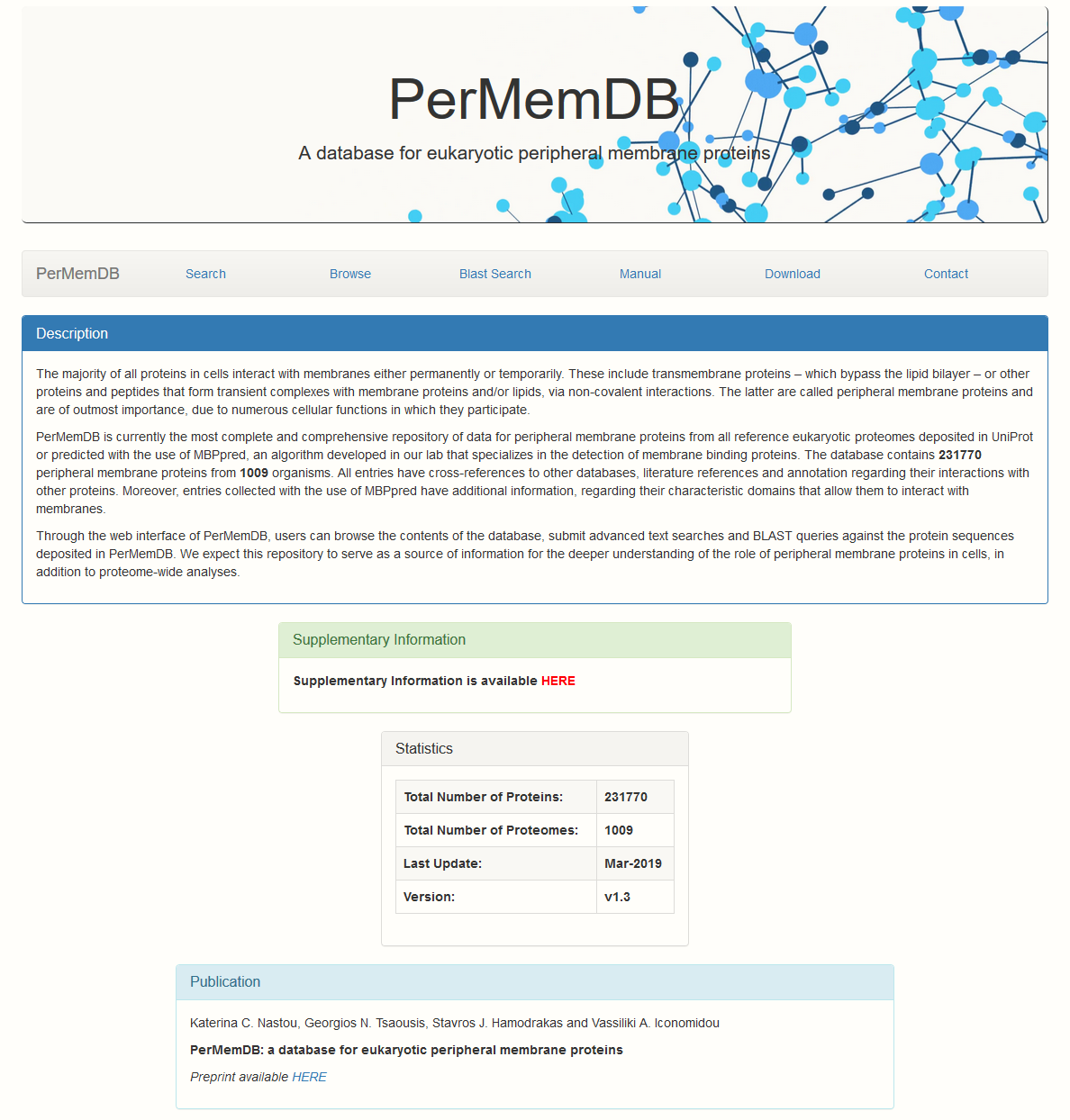
Home Page
In order to visit PerMemDB, the user should enter the following address:
http://bioinformatics.biol.uoa.gr/db=permemdb/.
The page loaded (see below) contains general information about the database and some statistics.

Search Page
In order to search database information, user should press the search button. A form appears with multiple options.
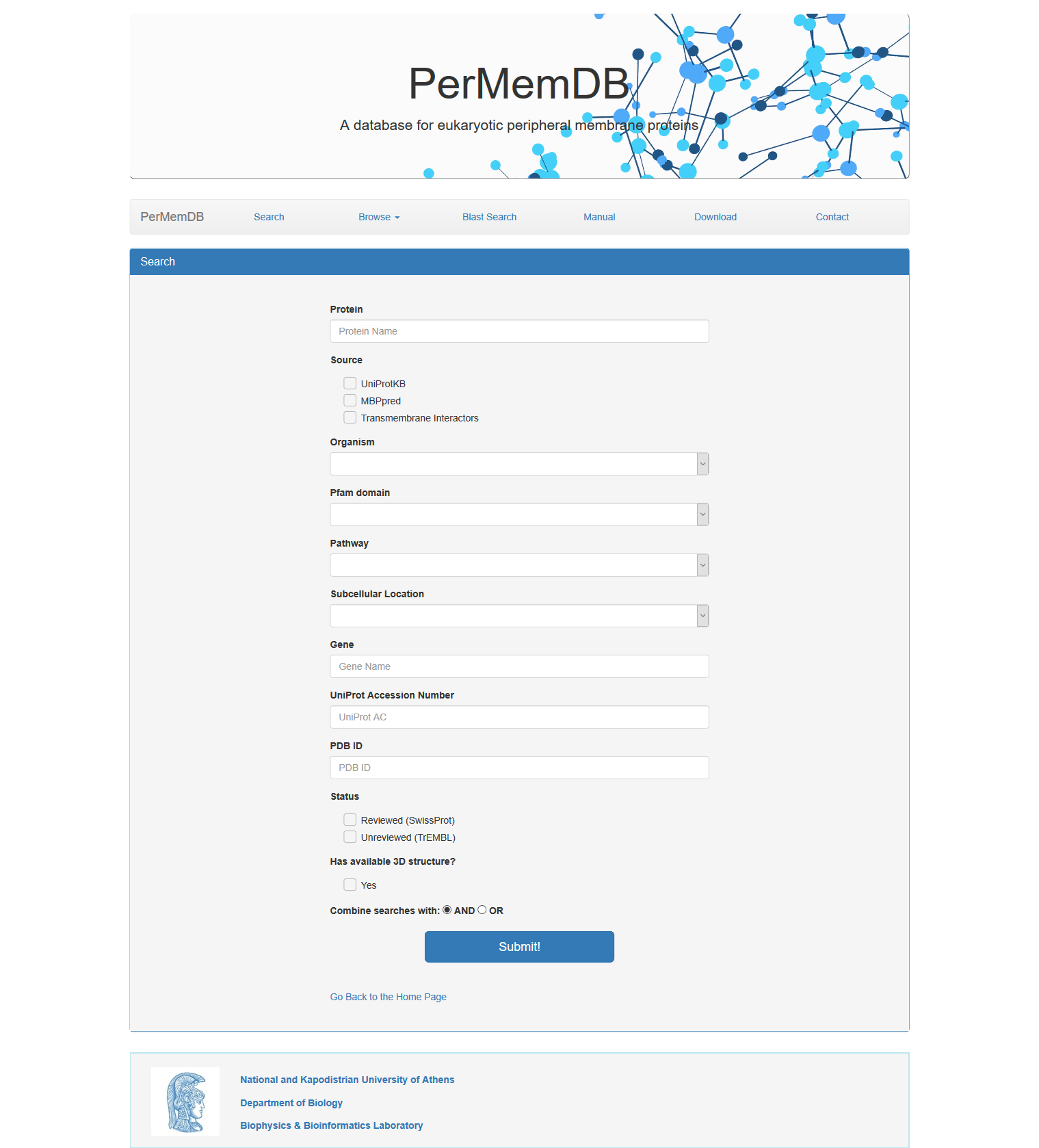 The choices are:
The choices are:
When a user selects a specific source of data, all entries will be shown, and when the sources are more than one (e.g. Transmembrane Interactor and MBPpred), the result table will list all sources.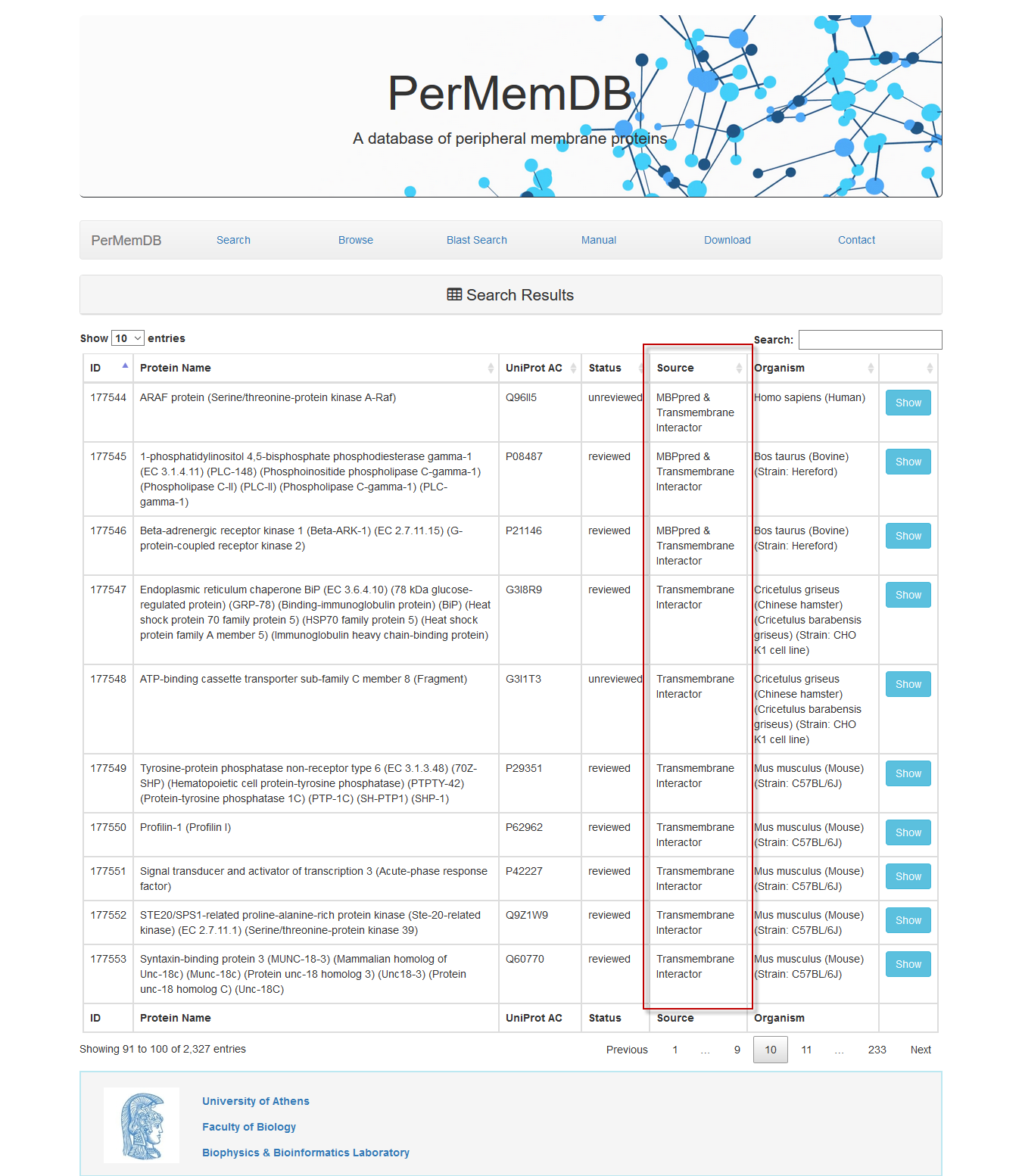
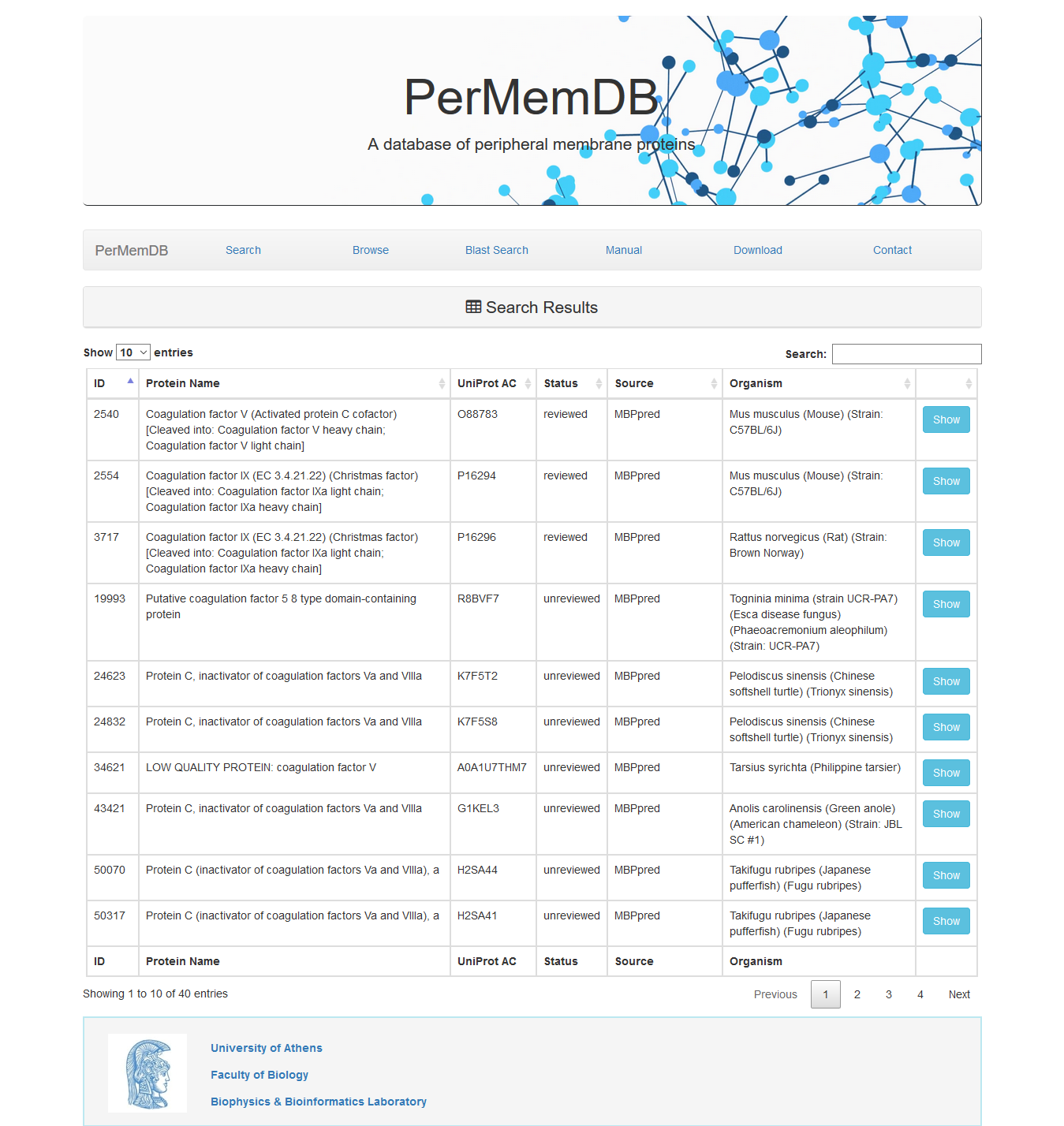
The search based on protein name and gene name does not require specific words. For example if a user enters the words 'coagulation factor' in the protein search field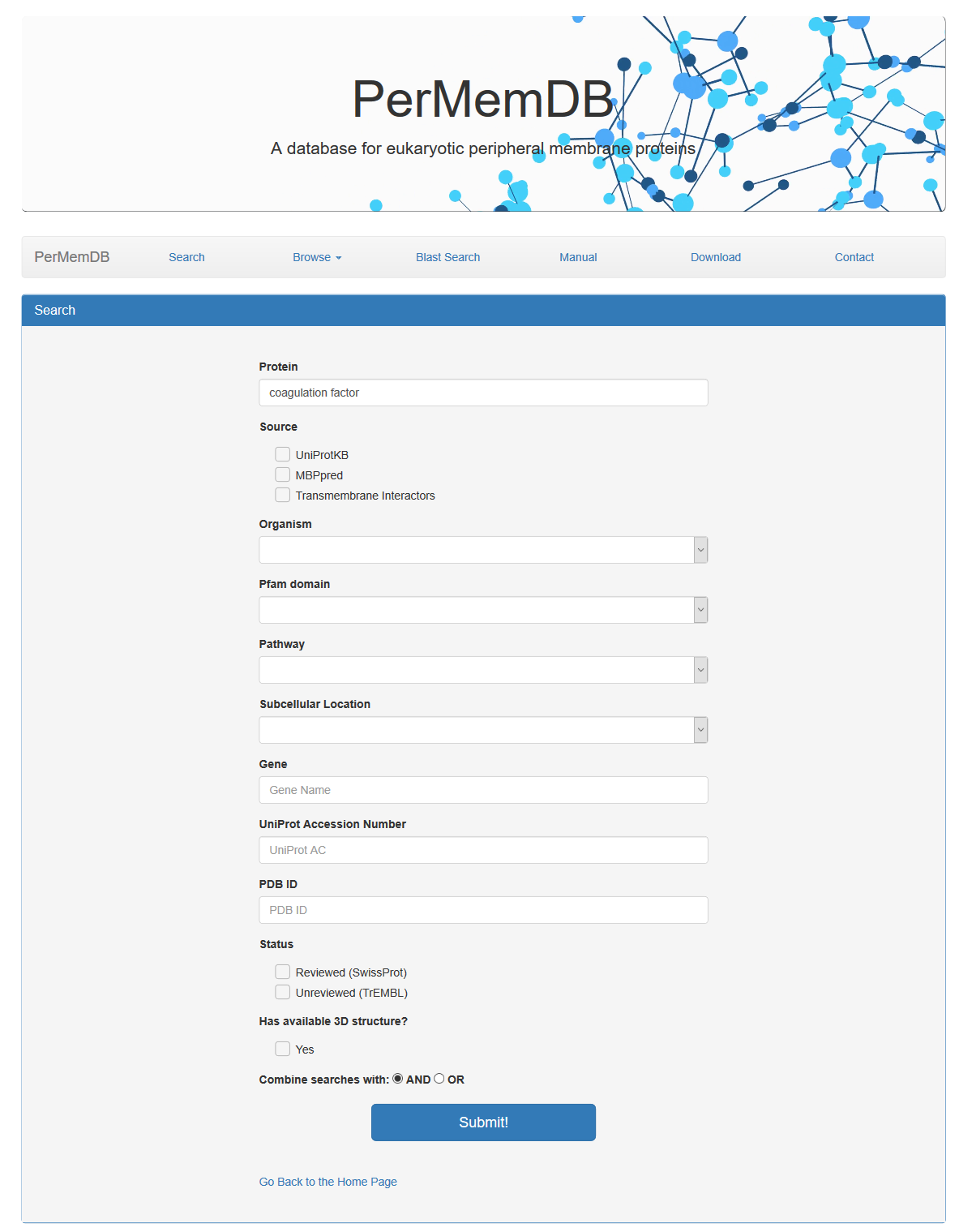 the result is all proteins whose name contains the words 'coagulation factor'.
the result is all proteins whose name contains the words 'coagulation factor'.
All searches can be combined with logical operators (AND/OR), in order to make the search result as specific as possible.
For example if we make the following combined search: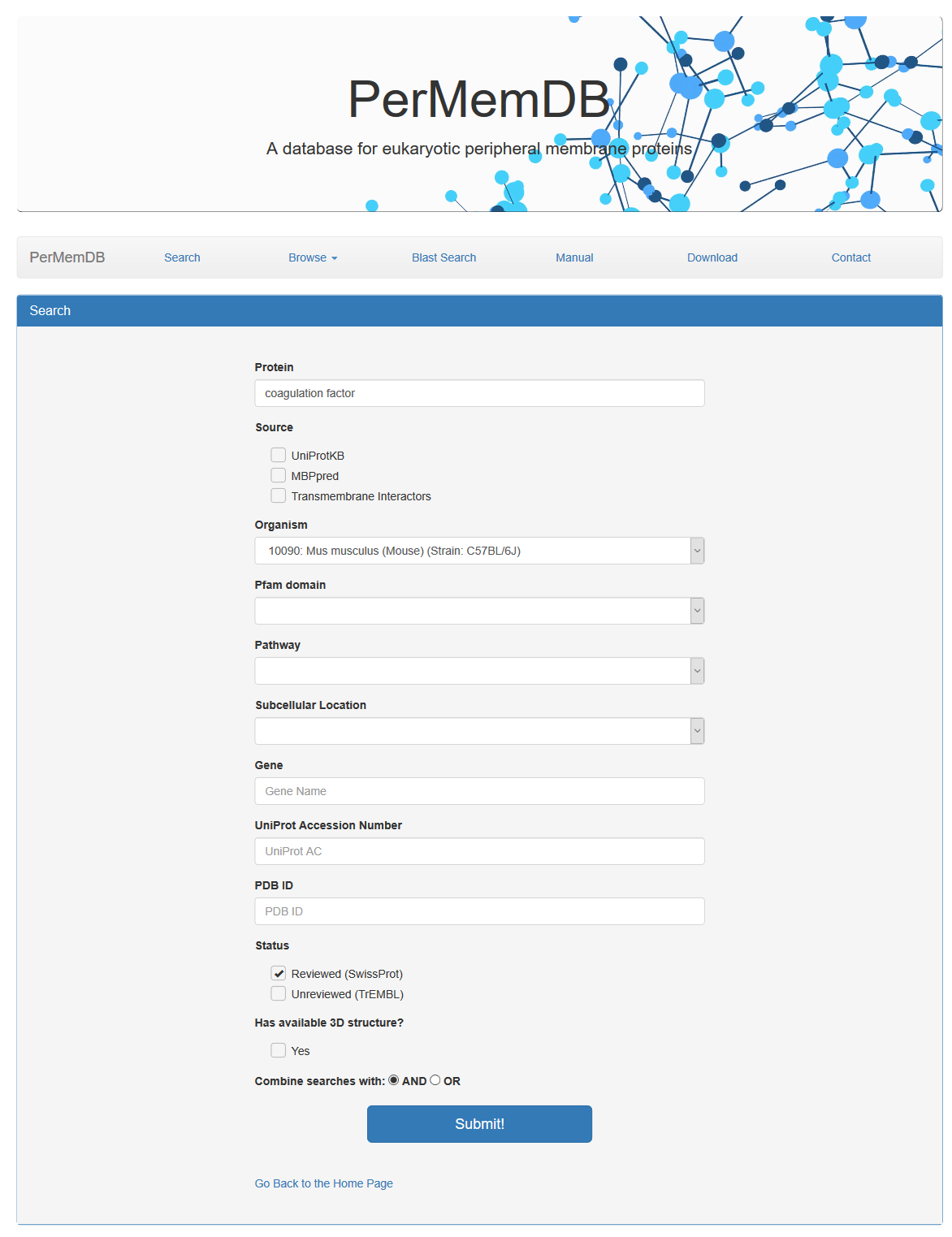 We get only two proteins with the specific characteristics.
We get only two proteins with the specific characteristics.
 The choices are:
The choices are:
- Search by Peripheral Membrane Protein Name
- Search by Source of Data. The choices are:
- UniProtKB
- MBPpred
- Transmembrane Interactors
- Search Proteins by Organism
- Search Proteins by Pfam domain
- Search Proteins by Pathway
- Search Proteins by Subcellular Location
- Search Proteins based on Gene Name
- Search Proteins based on Uniprot Accession or Uniprot Identifier
- Search Proteins based on PDB ID
- Search Proteins based on Protein Status (reviewed or unreviewed)
- Search Proteins based on 3D structure availability
When a user selects a specific source of data, all entries will be shown, and when the sources are more than one (e.g. Transmembrane Interactor and MBPpred), the result table will list all sources.

The search based on protein name and gene name does not require specific words. For example if a user enters the words 'coagulation factor' in the protein search field
 the result is all proteins whose name contains the words 'coagulation factor'.
the result is all proteins whose name contains the words 'coagulation factor'.

All searches can be combined with logical operators (AND/OR), in order to make the search result as specific as possible.
For example if we make the following combined search:
 We get only two proteins with the specific characteristics.
We get only two proteins with the specific characteristics.

Browse
Users can browse data using four options.
 The first option is to browse data by specific organism by pressing the respective button.
The first option is to browse data by specific organism by pressing the respective button.
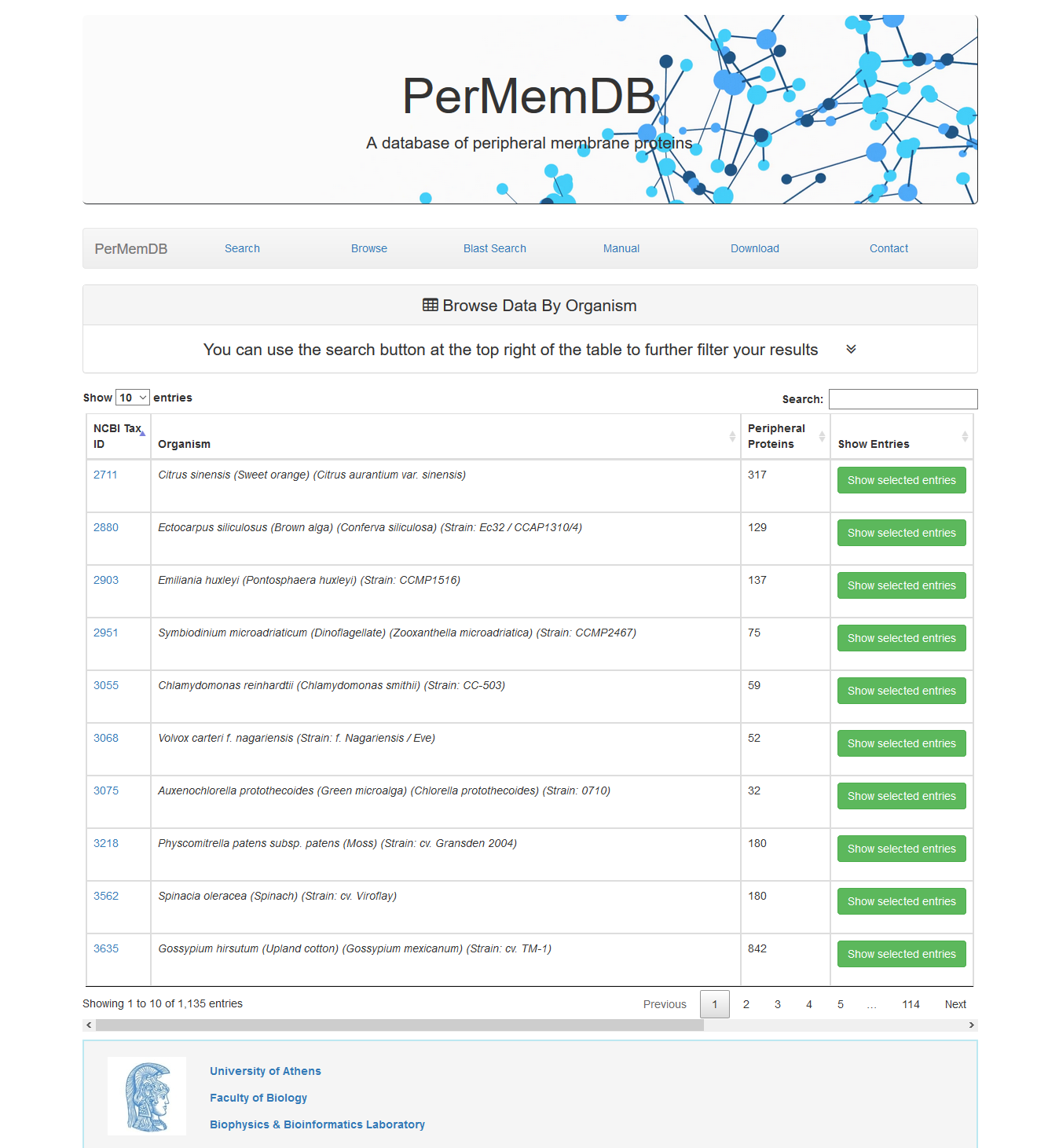
At first all entries appear. Each page shows 10 entries by default but this can be altered from the dropdown menu at the top left corner of the table to 25, 50 or all entries.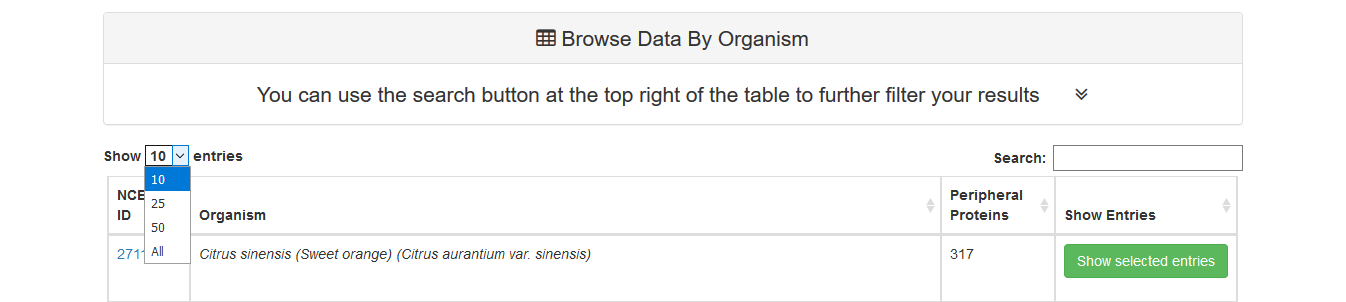
Moreover, users can perform non-specific searches using the search option at the top right corner of the data table. For example if the user searches for 'mouse' on the search field, all organisms with the word 'mouse' in their name will be shown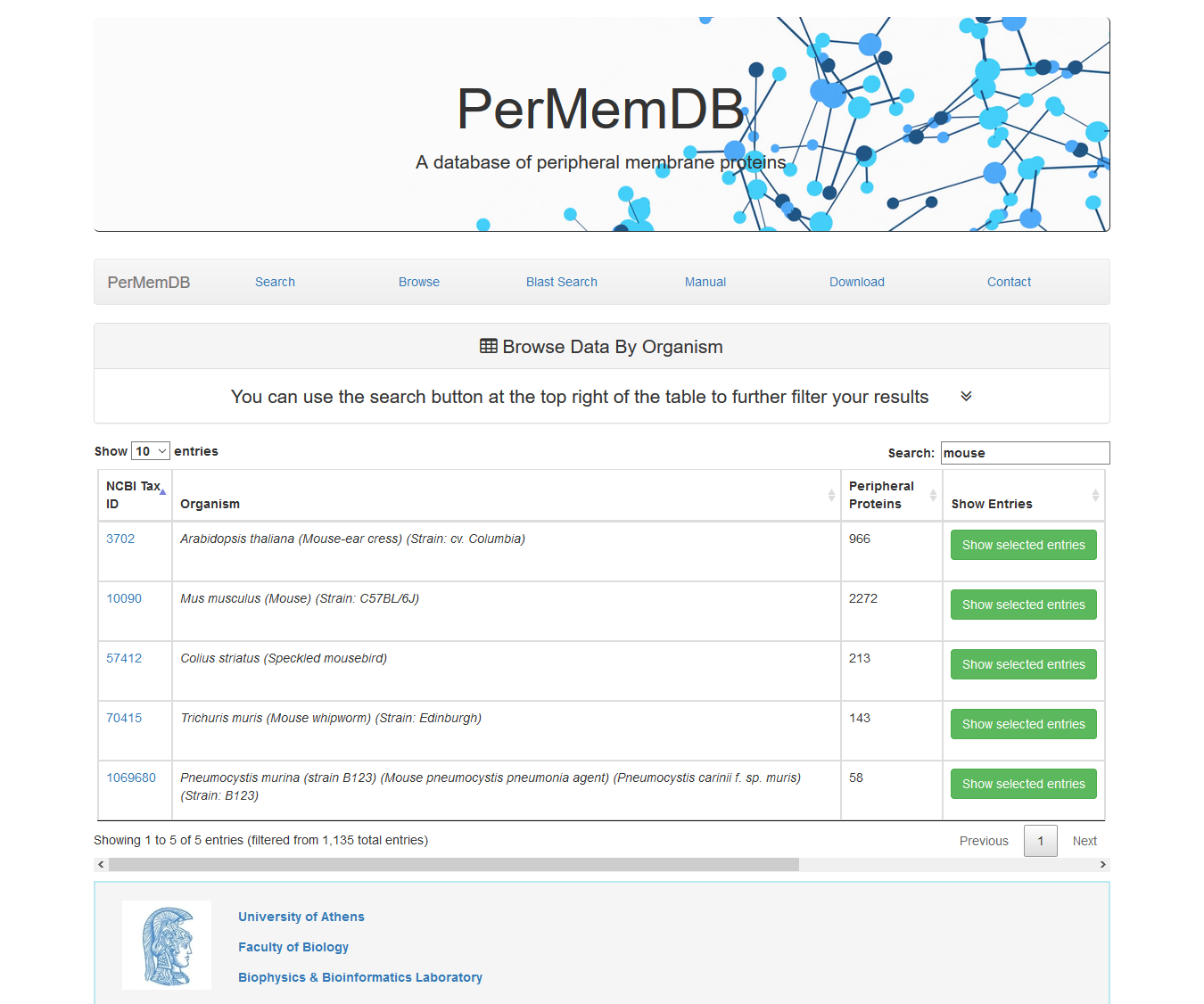 If a user presses Show Selected Entries all proteins from the selected organism are shown.
If a user presses Show Selected Entries all proteins from the selected organism are shown.
The second option is to browse data by Pfam domain.
The third option is to browse data by Pathway.
The fourth option is to browse data by Subcellular Location.
A drop-down menu appears for all three of the abovementioned options and a specific term can be selected.
e.g. A search for proteins that contain in their subcellular location field the entity "SL-9904: Single-pass membrane protein"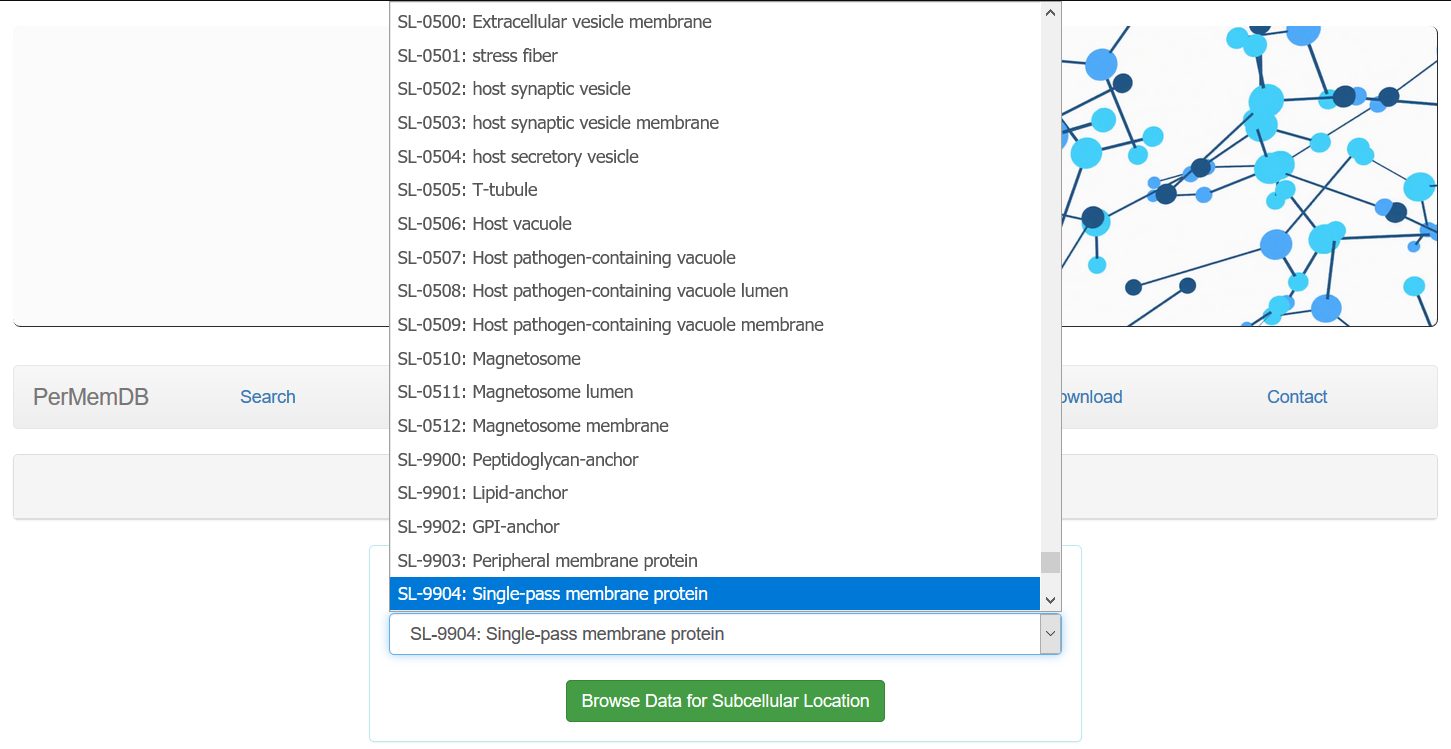 would result in 905 matching results.
would result in 905 matching results.
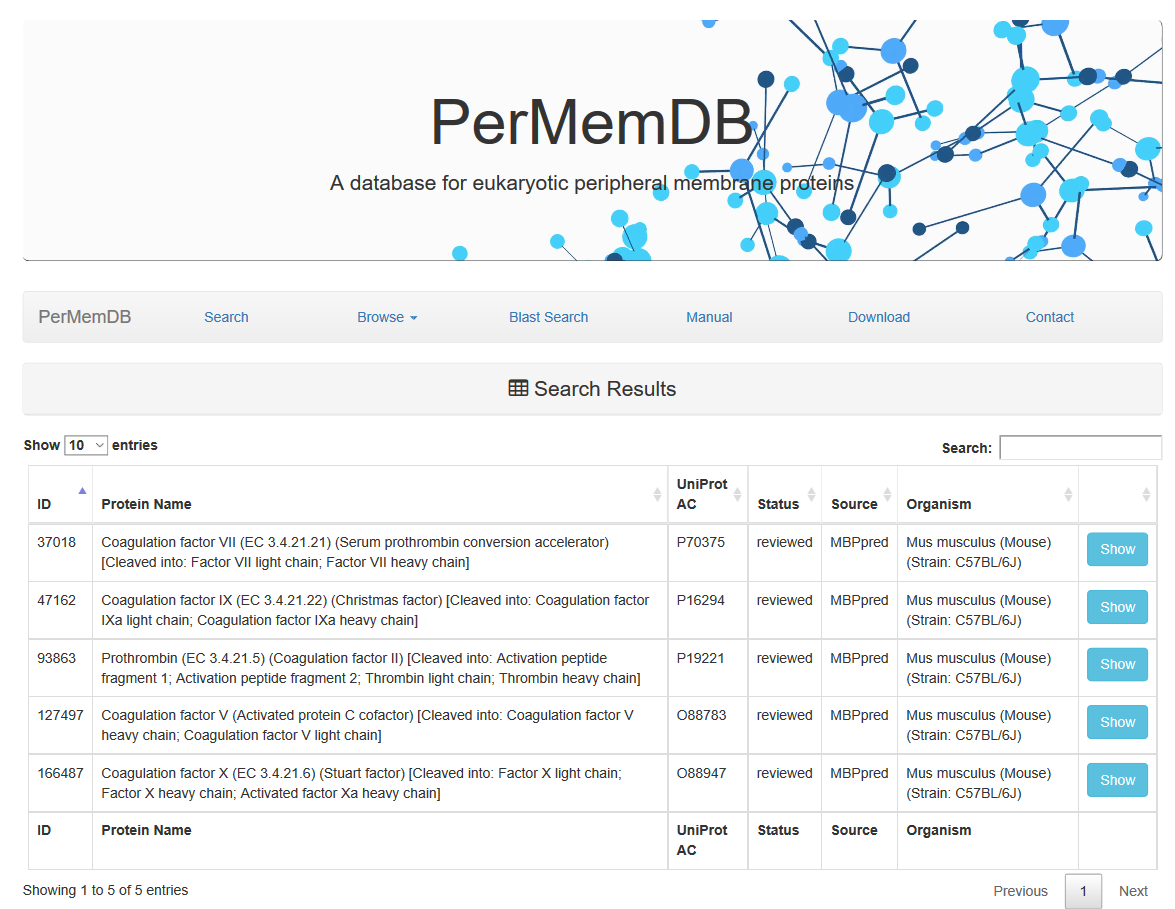
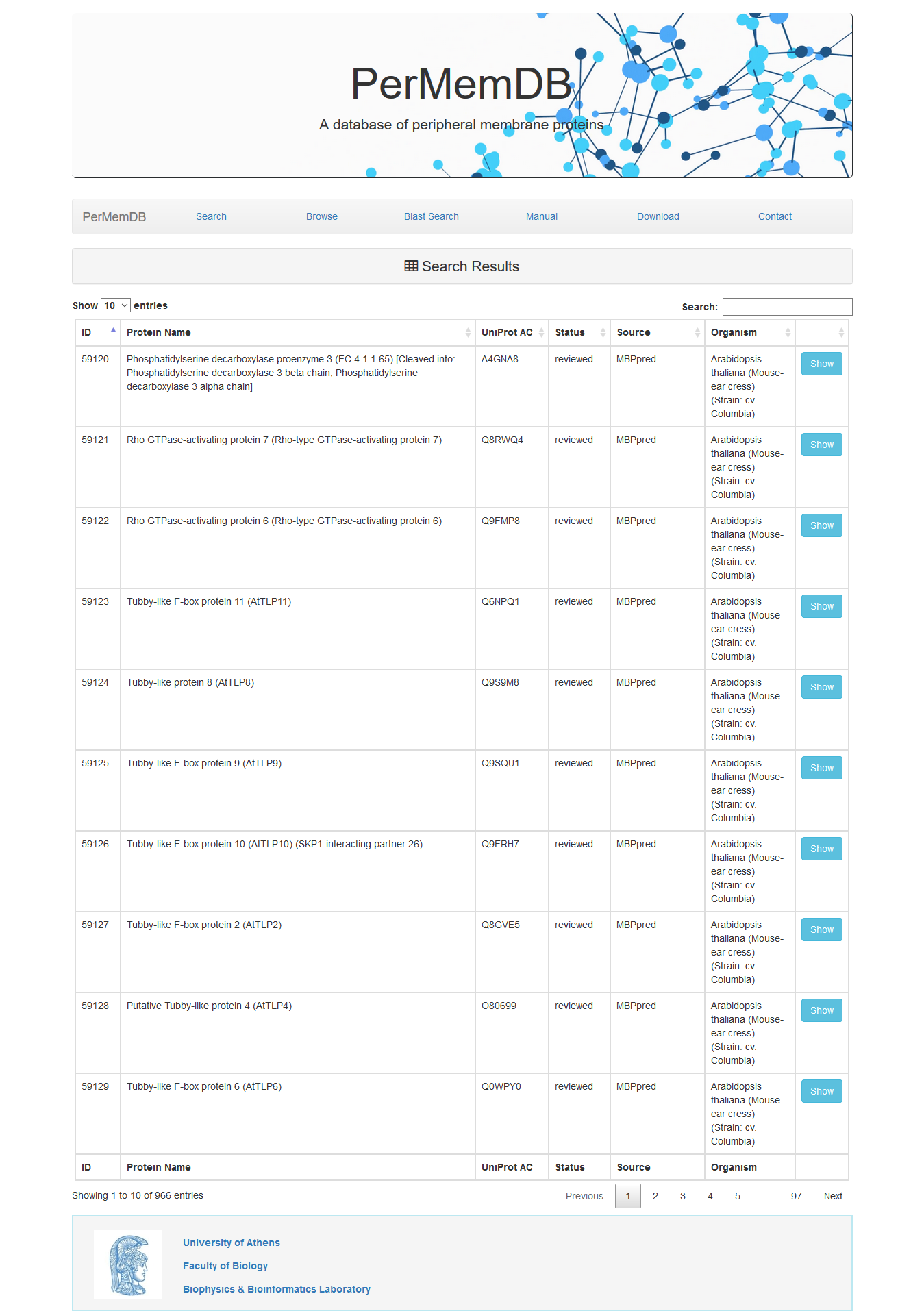
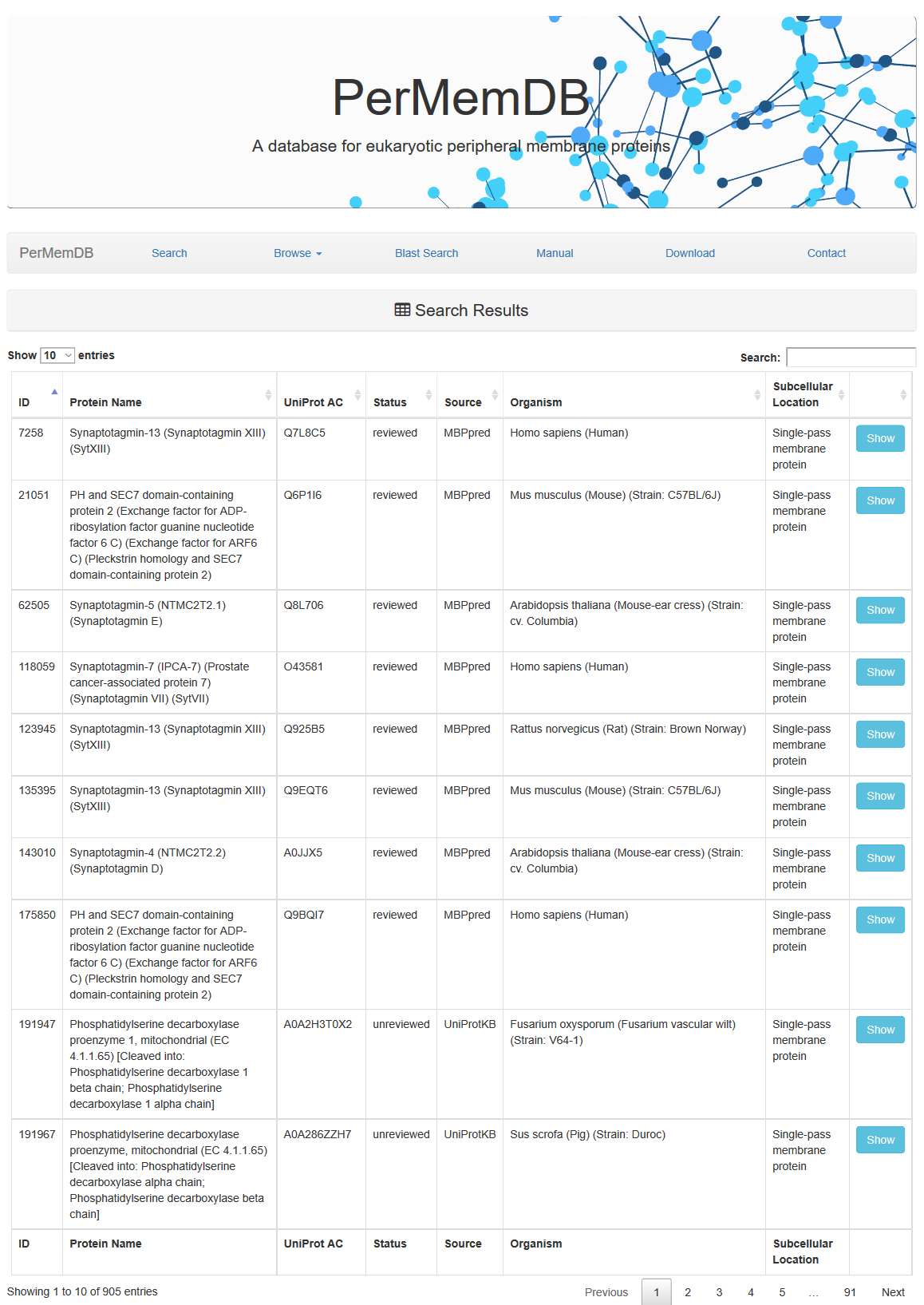 Through browsing the database or after a search is submitted, a list of proteins appears always,
as the one shown above. It contains the Protein Name, UniProt Accession, Status, Source of data and Organism.
Through browsing the database or after a search is submitted, a list of proteins appears always,
as the one shown above. It contains the Protein Name, UniProt Accession, Status, Source of data and Organism.
 The first option is to browse data by specific organism by pressing the respective button.
The first option is to browse data by specific organism by pressing the respective button.

At first all entries appear. Each page shows 10 entries by default but this can be altered from the dropdown menu at the top left corner of the table to 25, 50 or all entries.

Moreover, users can perform non-specific searches using the search option at the top right corner of the data table. For example if the user searches for 'mouse' on the search field, all organisms with the word 'mouse' in their name will be shown
 If a user presses Show Selected Entries all proteins from the selected organism are shown.
If a user presses Show Selected Entries all proteins from the selected organism are shown.

The second option is to browse data by Pfam domain.

The third option is to browse data by Pathway.

The fourth option is to browse data by Subcellular Location.

A drop-down menu appears for all three of the abovementioned options and a specific term can be selected.
e.g. A search for proteins that contain in their subcellular location field the entity "SL-9904: Single-pass membrane protein"
 would result in 905 matching results.
would result in 905 matching results.
 Through browsing the database or after a search is submitted, a list of proteins appears always,
as the one shown above. It contains the Protein Name, UniProt Accession, Status, Source of data and Organism.
Through browsing the database or after a search is submitted, a list of proteins appears always,
as the one shown above. It contains the Protein Name, UniProt Accession, Status, Source of data and Organism.
Entry
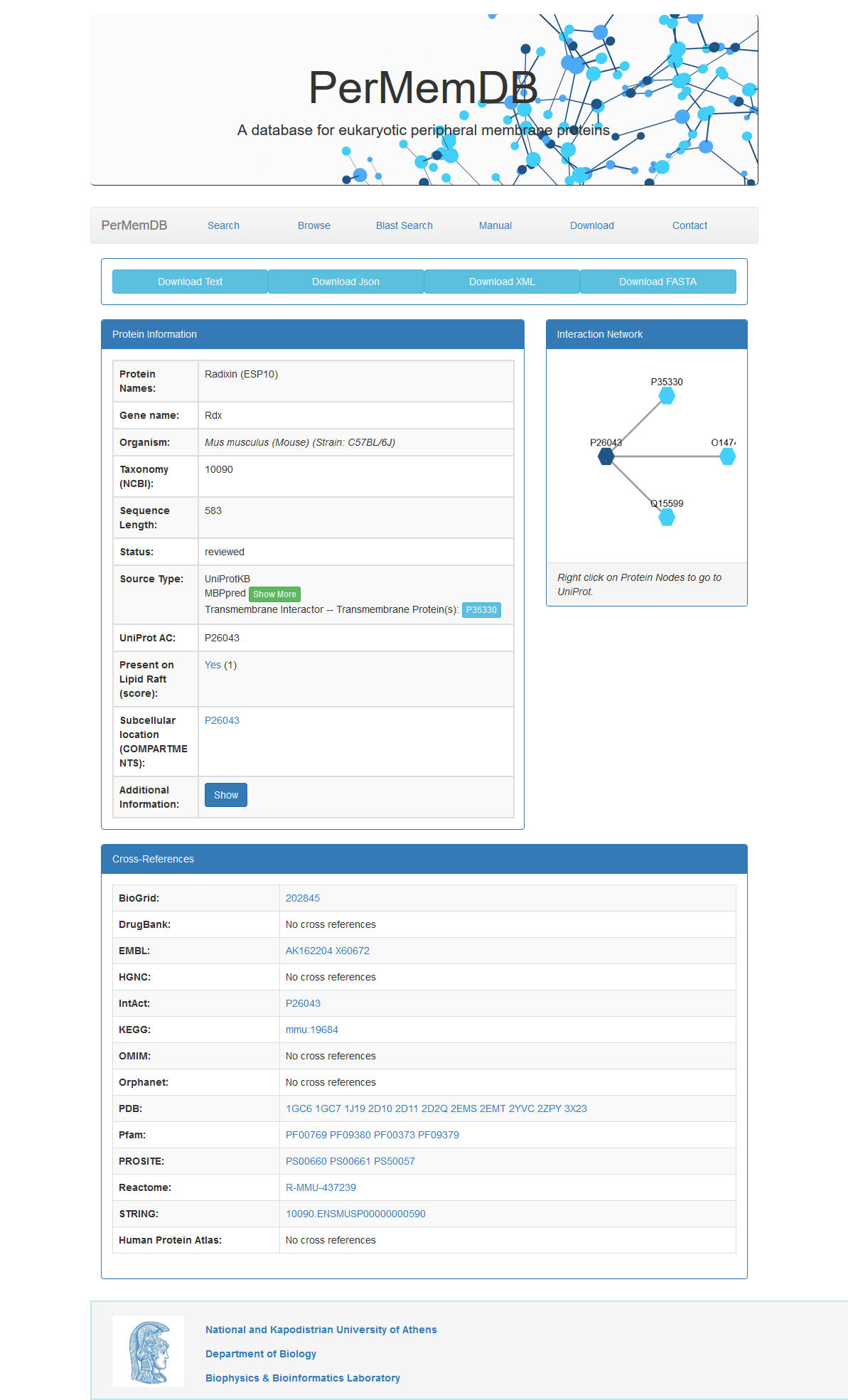
When the user presses the Show button they are redirected to the Entry page.
The Entry page contains information about the protein as explained below.
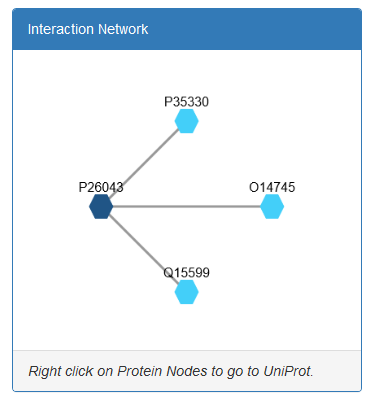
Moreover Cytoscape JS is integrated to visualize the relationship between
peripheral proteins and their interactors.
 More specifically:
More specifically:
 More specifically:
More specifically:
-
In the top of the page the user can find four buttons.
Download Text, Download Json, Download XML and Download FASTA.
By pressing these buttons the user can download entry information in text, Json, XML or FASTA format respectively.

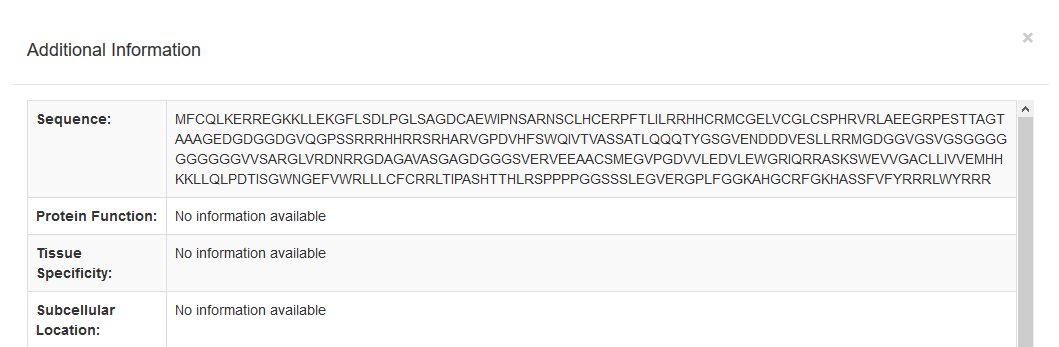
- The basic protein information available is:
- Protein Name
- Gene Name
- Organism
- NCBI Taxonomic ID
- Sequence Length
- Status
- Source Type(s)
- UniProt AC
- Presence on Lipid Raft (Data from RaftProt)
- Subcellular Localization (Data from COMPARTMENTS)
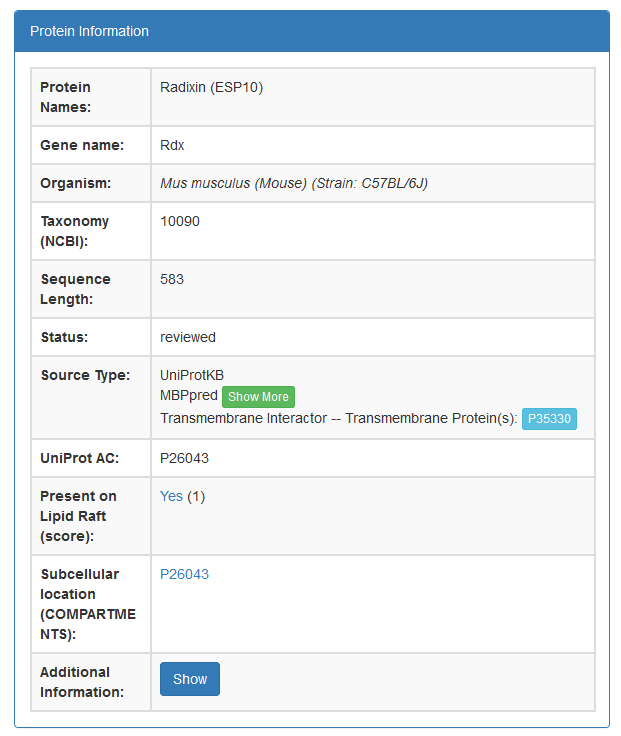
- Additional information for the protein's sequence, function, tissue specificity and subcellular location can be found when a user presses the blue show button
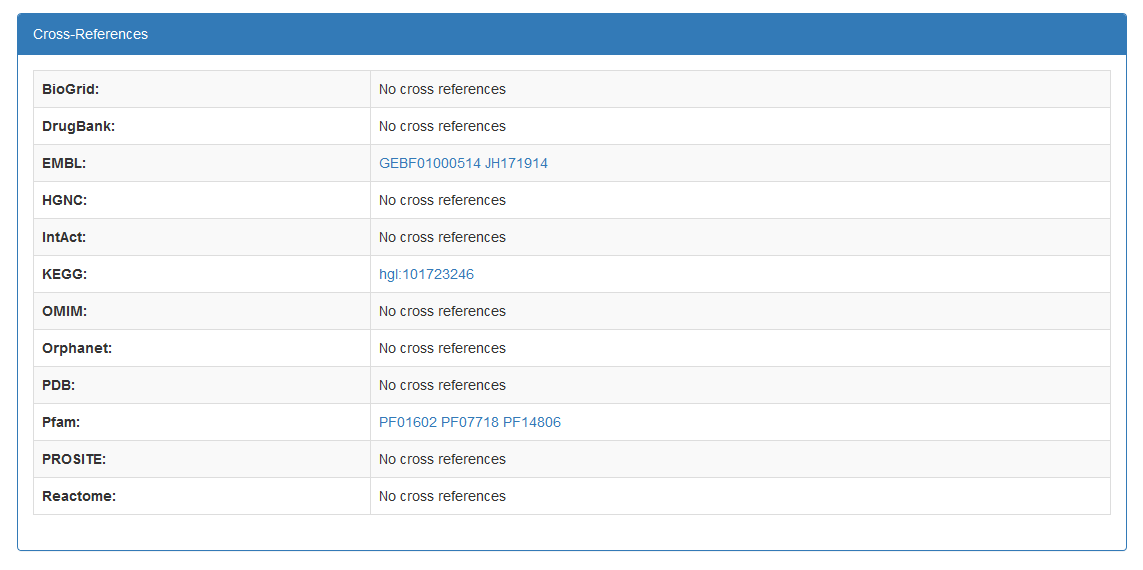
- The panel at the bottom of the page contains references to other databases. The databases are:
- BioGrid
- DrugBank
- EMBL
- HGNC
- IntAct
- KEGG
- OMIM
- Orphanet
- PDB
- Pfam
- PROSITE
- Reactome
- STRING
- HPA
-
When the user presses on the green Show More button next to MBPpred, information produced by the algorithm are shown.
These information include:
- Domain Family Name (as Shown in Pfam)
- Domain Position and Score
- Domain on the Protein Sequence (red)
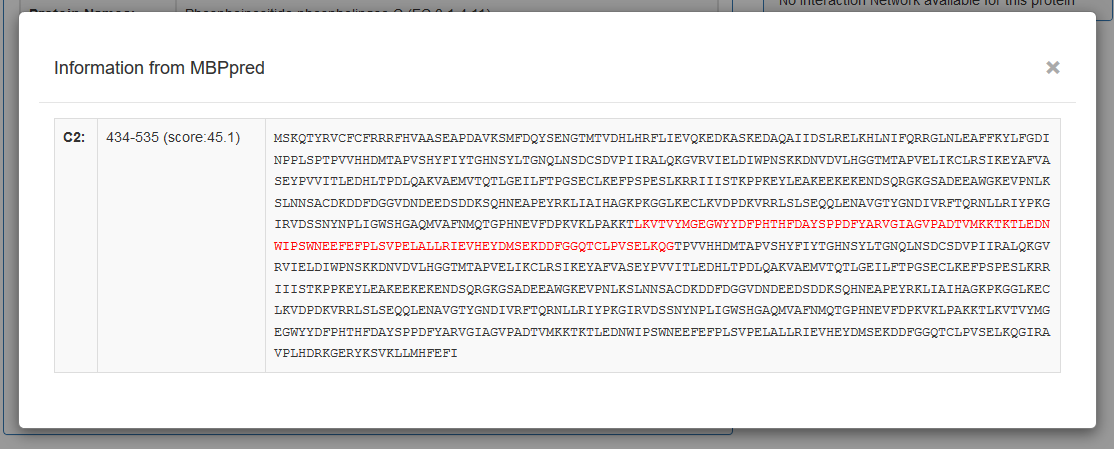
- Finally, a Cytoscape JS viewer is integrated in the page for vizualization of undirected graphs showing the interactions between each peripheral membrane protein (dark blue) with other proteins (light blue). The user can right click on protein nodes and get redirected to UniProt.
BLAST search
With the BLAST search tool, the user may submit a sequence and search the database for finding homologues.
The input for the BLAST application is the sequence in standard FASTA format.
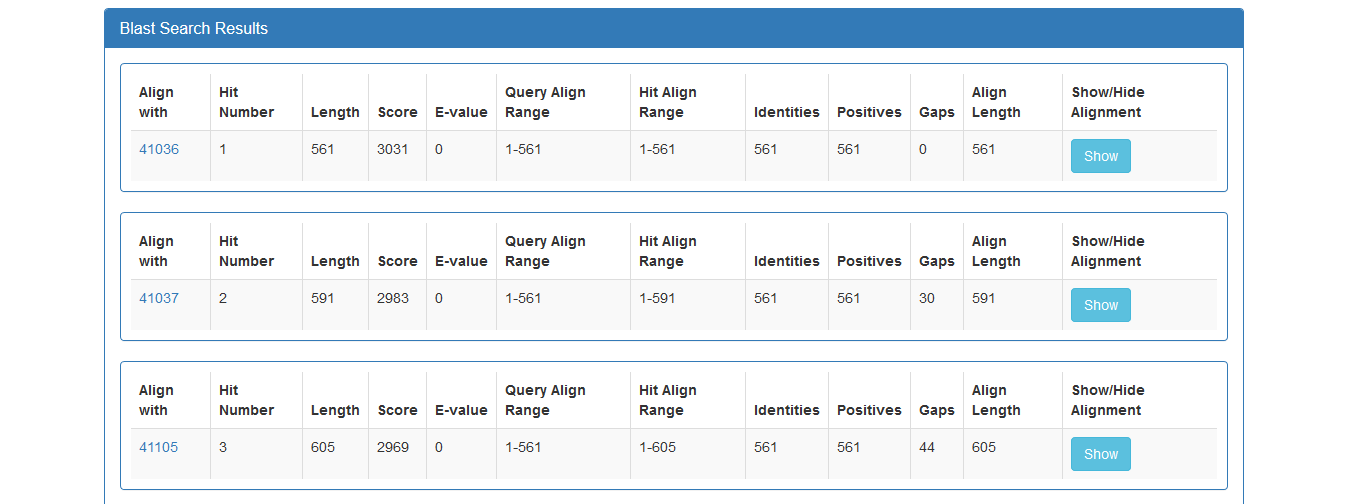
 The result page of the BLAST search shows a list of the Blast hits with significant alignment
on the query sequence the user has submitted. The list is in a table format including the
target protein, the Length of the target sequence and the Query and Target
align range. The BLAST results can be compared through the Score and E-value and the Identities and Positives.
The result page of the above BLAST search is:
The result page of the BLAST search shows a list of the Blast hits with significant alignment
on the query sequence the user has submitted. The list is in a table format including the
target protein, the Length of the target sequence and the Query and Target
align range. The BLAST results can be compared through the Score and E-value and the Identities and Positives.
The result page of the above BLAST search is:
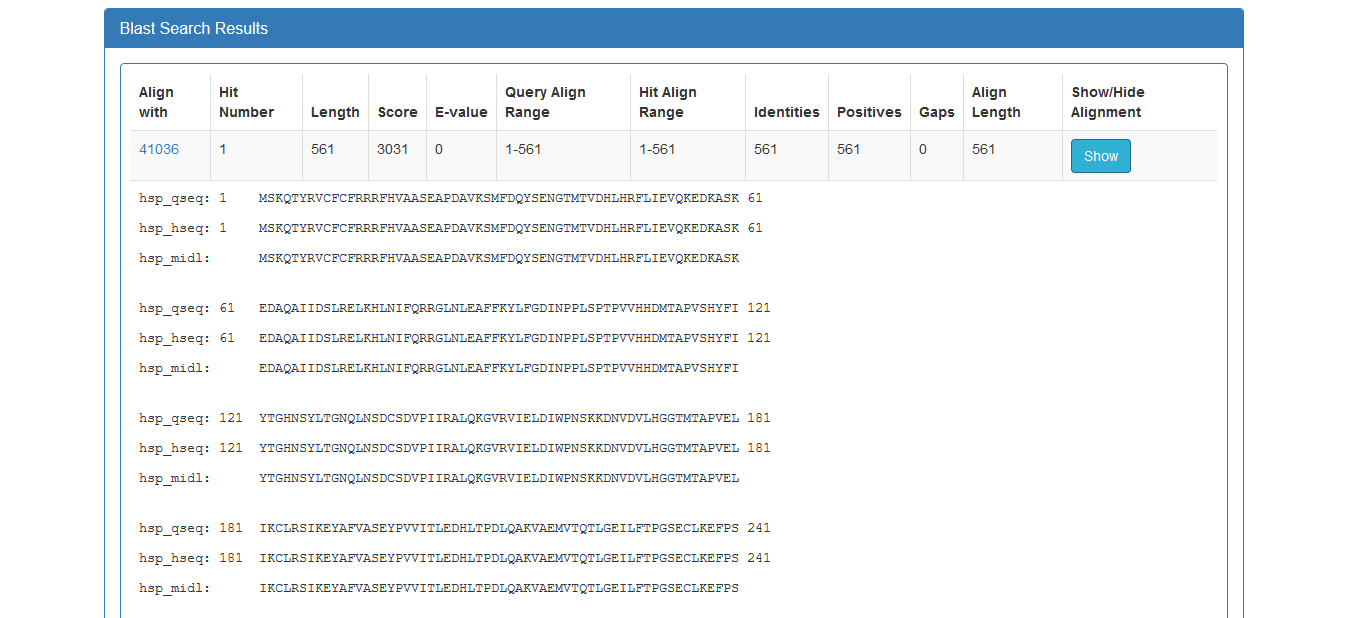
 Furthermore, the user can have a more detailed view of each alignment through the Show button at the end of each line:
Furthermore, the user can have a more detailed view of each alignment through the Show button at the end of each line:
 The result page of the BLAST search shows a list of the Blast hits with significant alignment
on the query sequence the user has submitted. The list is in a table format including the
target protein, the Length of the target sequence and the Query and Target
align range. The BLAST results can be compared through the Score and E-value and the Identities and Positives.
The result page of the above BLAST search is:
The result page of the BLAST search shows a list of the Blast hits with significant alignment
on the query sequence the user has submitted. The list is in a table format including the
target protein, the Length of the target sequence and the Query and Target
align range. The BLAST results can be compared through the Score and E-value and the Identities and Positives.
The result page of the above BLAST search is:
 Furthermore, the user can have a more detailed view of each alignment through the Show button at the end of each line:
Furthermore, the user can have a more detailed view of each alignment through the Show button at the end of each line:

Download Page
User can download all database files in Text, JSON, XML or FASTA format.
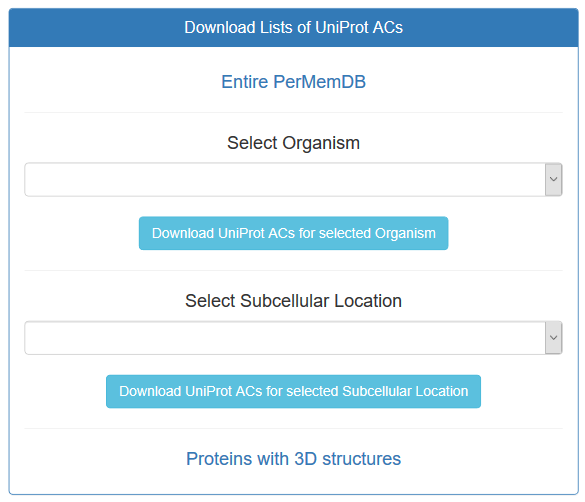
 Moreover, lists of UniProt ACs are provided for all entries of the database, entries with 3D structures, and all entries belonging to different organisms represented in the database or located on different subcellular locations.
Moreover, lists of UniProt ACs are provided for all entries of the database, entries with 3D structures, and all entries belonging to different organisms represented in the database or located on different subcellular locations.
 Moreover, lists of UniProt ACs are provided for all entries of the database, entries with 3D structures, and all entries belonging to different organisms represented in the database or located on different subcellular locations.
Moreover, lists of UniProt ACs are provided for all entries of the database, entries with 3D structures, and all entries belonging to different organisms represented in the database or located on different subcellular locations.

Contact Page
Users can contact us for more information at the emails specified at the contact page.
 If a user wants to submit data or corrections that will be reviewed and later will be added to the database
a form has been created to provide their name, email address and a message that will be
reviewed by the authors.
If a user wants to submit data or corrections that will be reviewed and later will be added to the database
a form has been created to provide their name, email address and a message that will be
reviewed by the authors.
 Related publications to the current work are also presented.
Related publications to the current work are also presented.
 If a user wants to submit data or corrections that will be reviewed and later will be added to the database
a form has been created to provide their name, email address and a message that will be
reviewed by the authors.
If a user wants to submit data or corrections that will be reviewed and later will be added to the database
a form has been created to provide their name, email address and a message that will be
reviewed by the authors.
 Related publications to the current work are also presented.
Related publications to the current work are also presented.
Technologies
PerMemDB is based on modern technologies. User should have Javascript enabled on the web browser.

